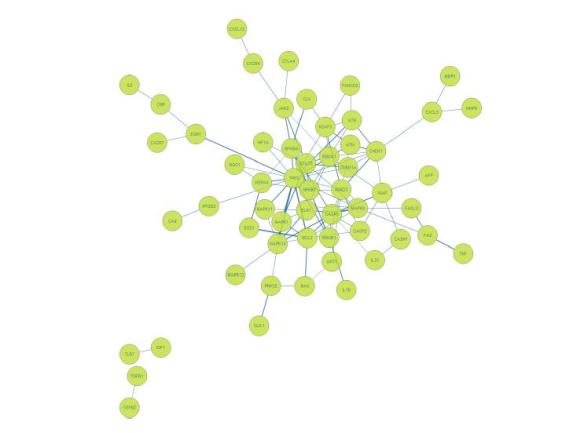
Clustering biological networks is extremely significant in the field of molecular biology and its applications as it allows for the functional annotation of cellular molecules and the prediction of physical complexes formulated by proteins and/or RNA molecules.
InSyBio’s scientific team has recently published a new methodology (Gradually Expanding Dense Neighborhoods – GENA) that encompasses several properties, such as handling effectively weighted biological networks and predicting overlapping clusters, which allow it to significantly outperform the state-of-the-art methods.
The Research and Development Manager of InSyBio Christos Dimitrakopoulos, who was the head of this work, has stated that “The computational prediction of protein complexes is becoming more precise and informative. GENA makes realistic assumptions about the underlying molecular mechanisms of the cell“.
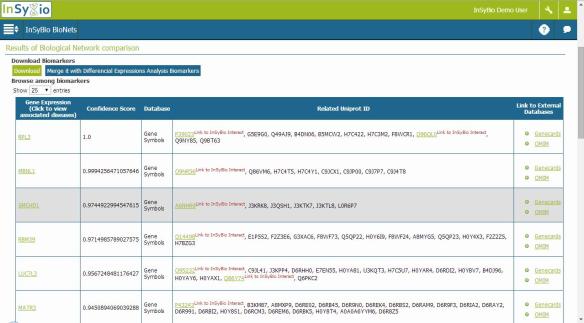
The full paper is published by the prestigious journal of Artificial Intelligence in Medicine with the title: “Predicting overlapping protein complexes from weighted protein interaction graphs by gradually expanding dense neighborhoods” and it is available online at http://dx.doi.org/10.1016/j.artmed.2016.05.006 . The implementation of the code is available upon request (requests should be sent at info@insybio.com) for academic usage only, while it will also be integrated in the next version of InSyBio BioNets tool, which is a web based tool integrated in InSyBio Suite (try it at http://demo.insybio.com).
About InsyBio: InSyBio is a bioinformatics pioneer company in personalized healthcare which focuses on developing computational frameworks and tools for the analysis of complex life-science and biological data. The key objective of our analysis lies in the discovery of predictive integrated biomarkers (biomarkers of various categories) with increased prognostic and diagnostic aspects for personalized Healthcare Industry. Find more details at www.insybio.com